A Moment in Time
Submitted by: Luke Shinneman
Category: Microscopy
Contributions: Leo Green
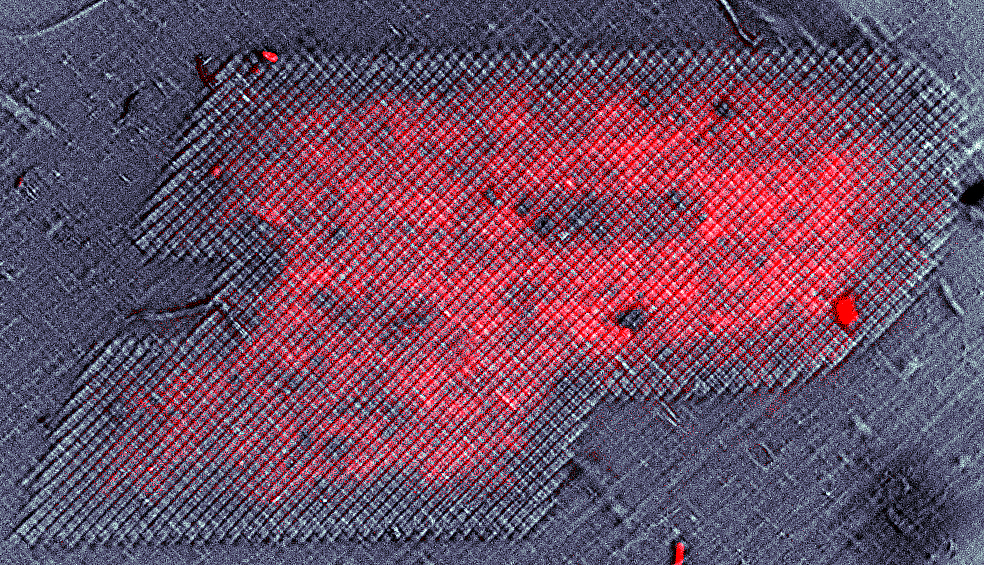
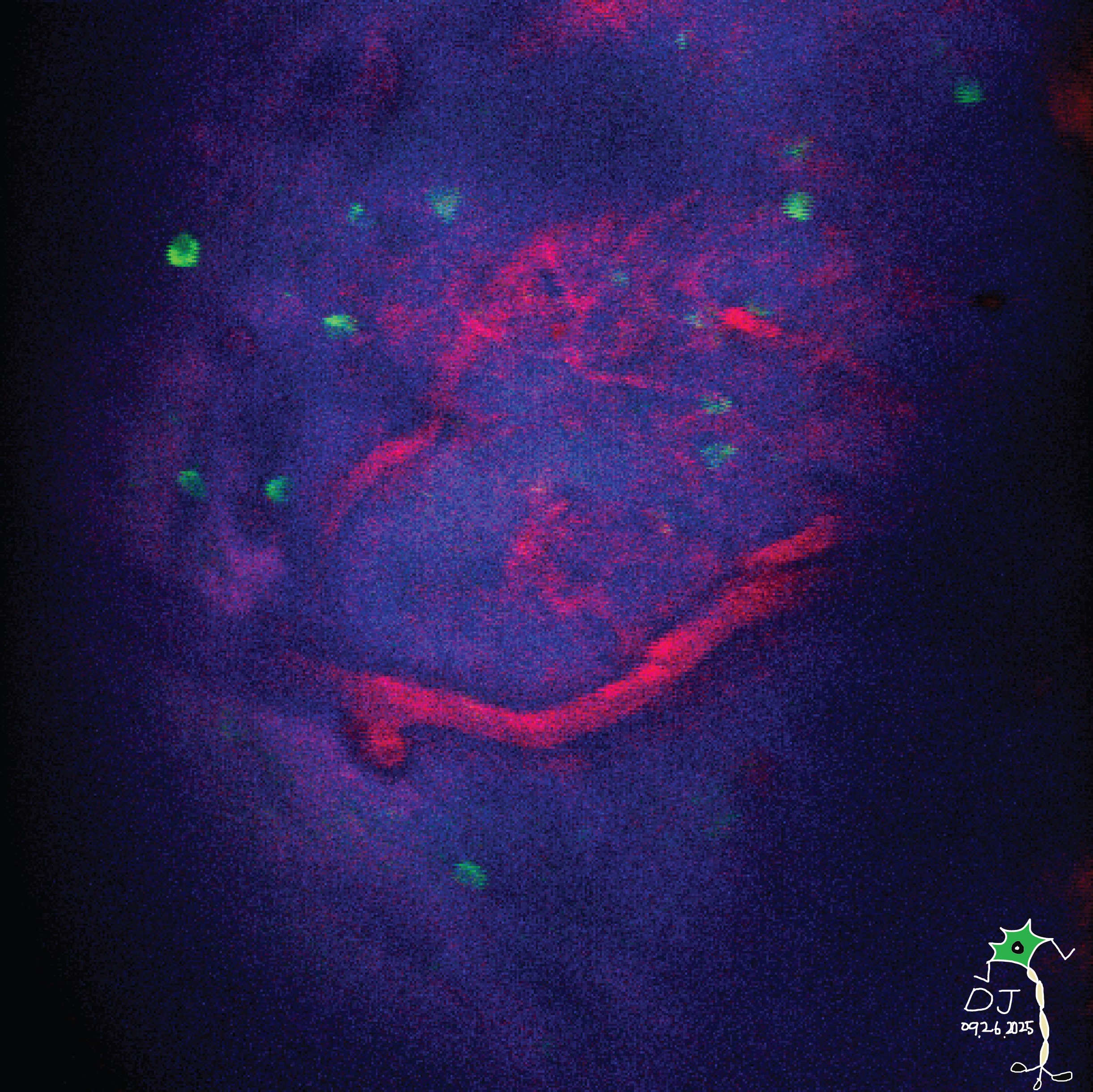
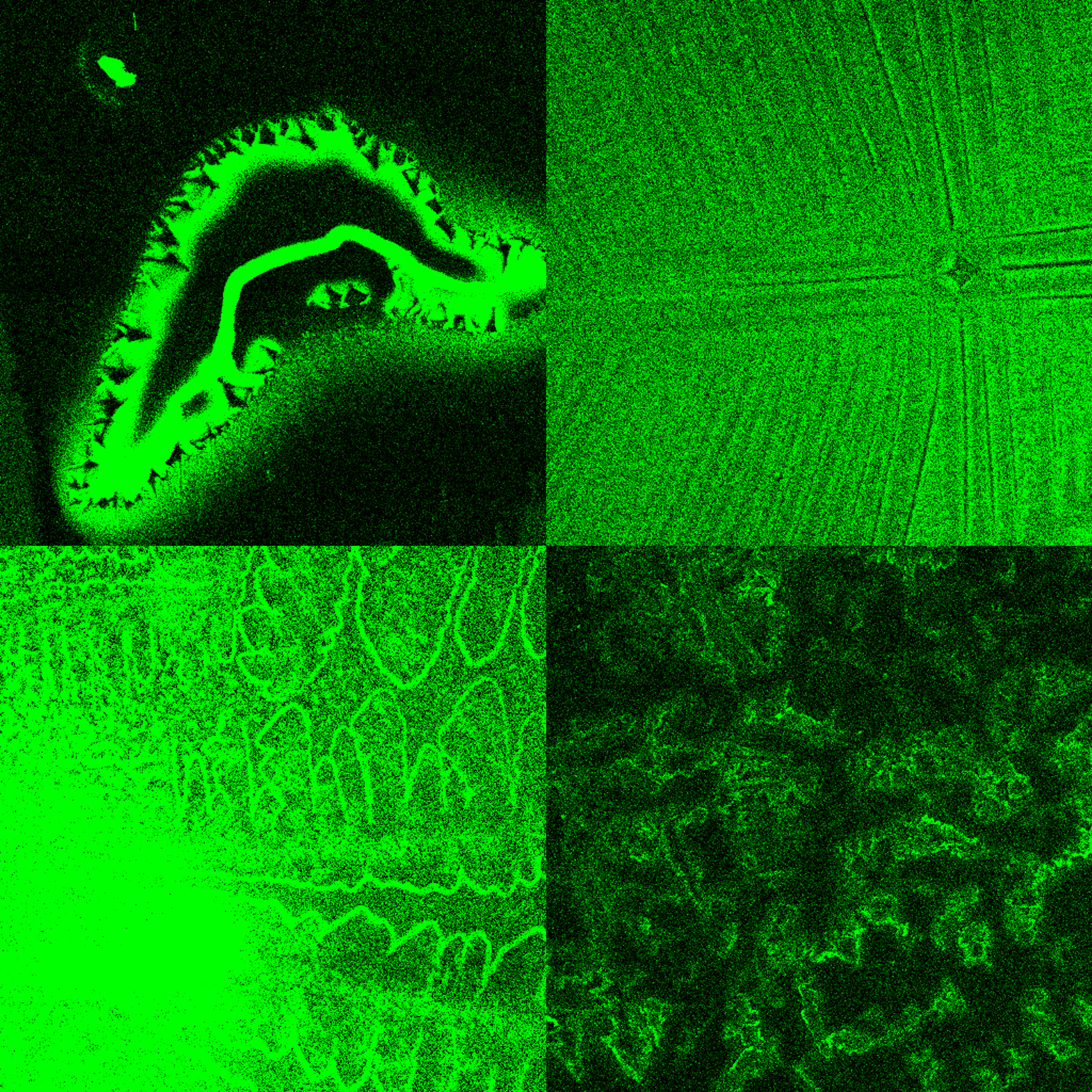
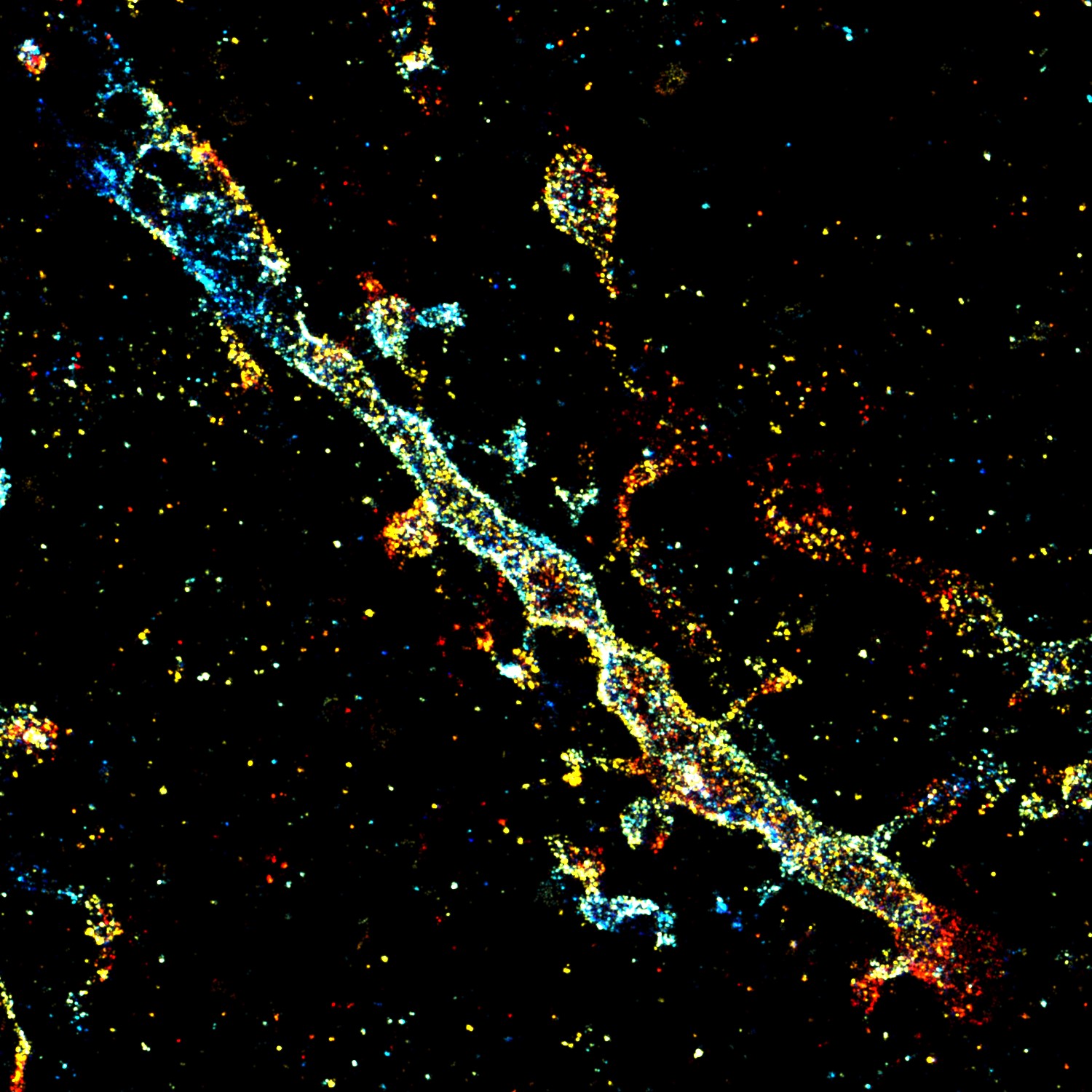
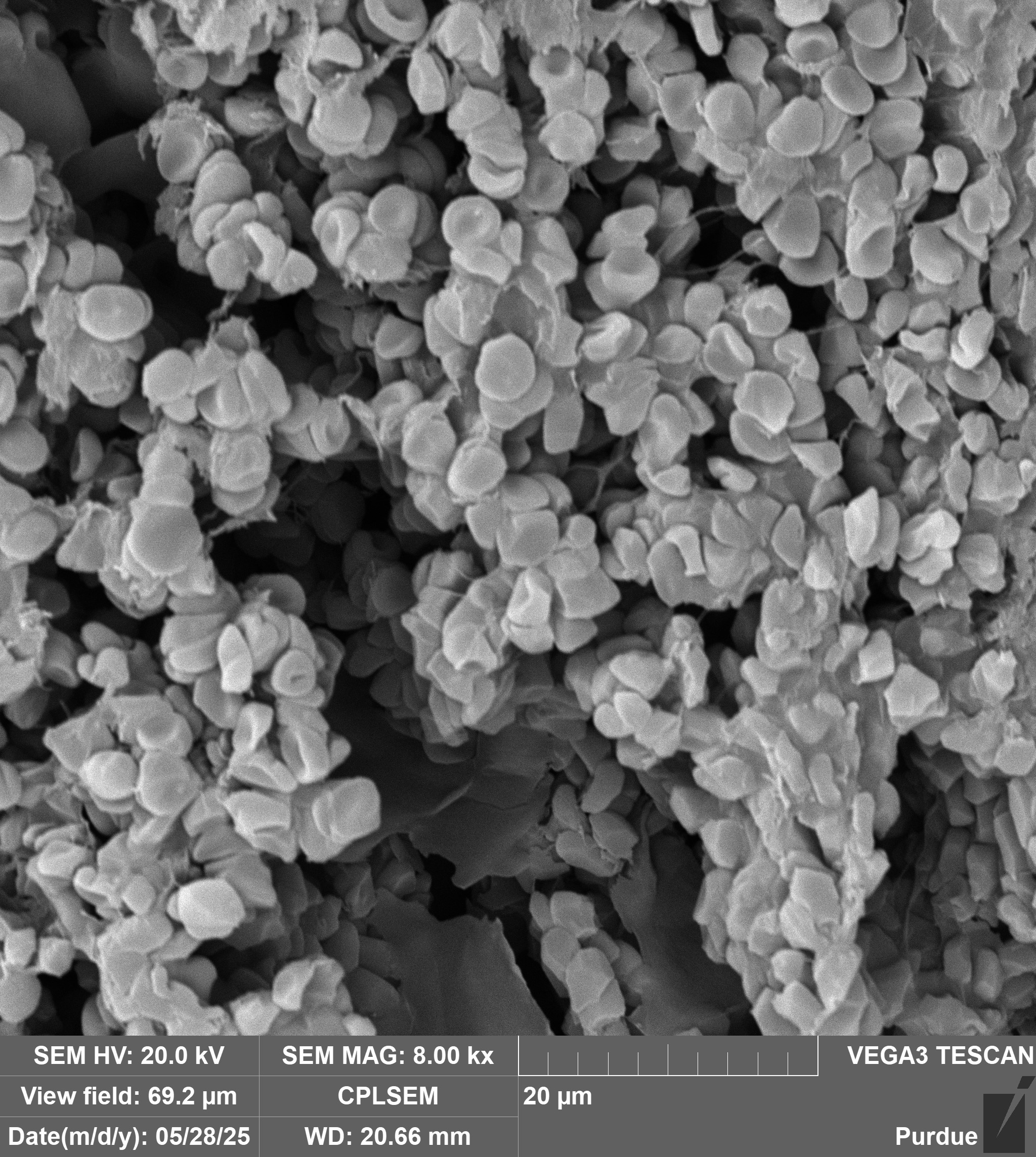
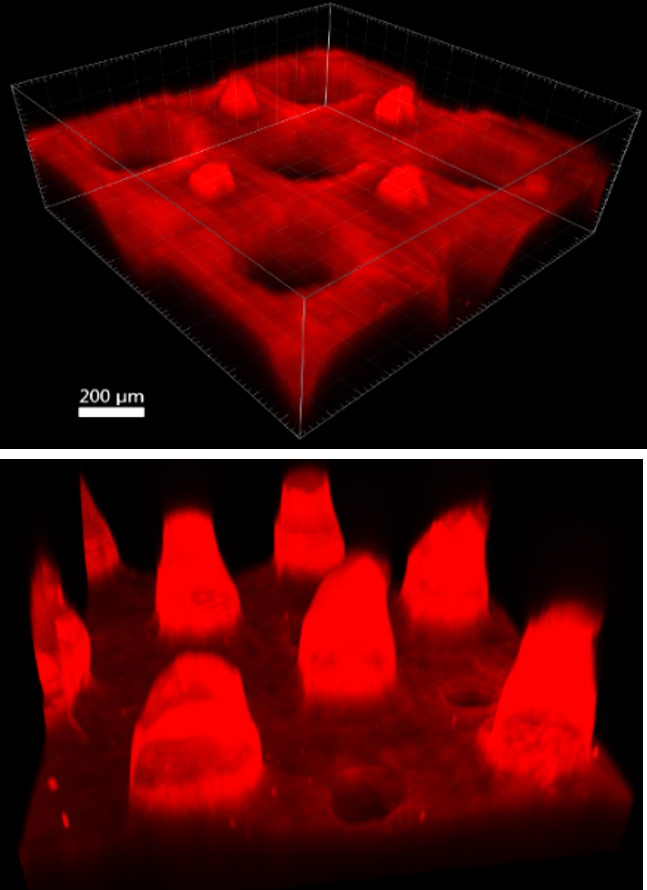
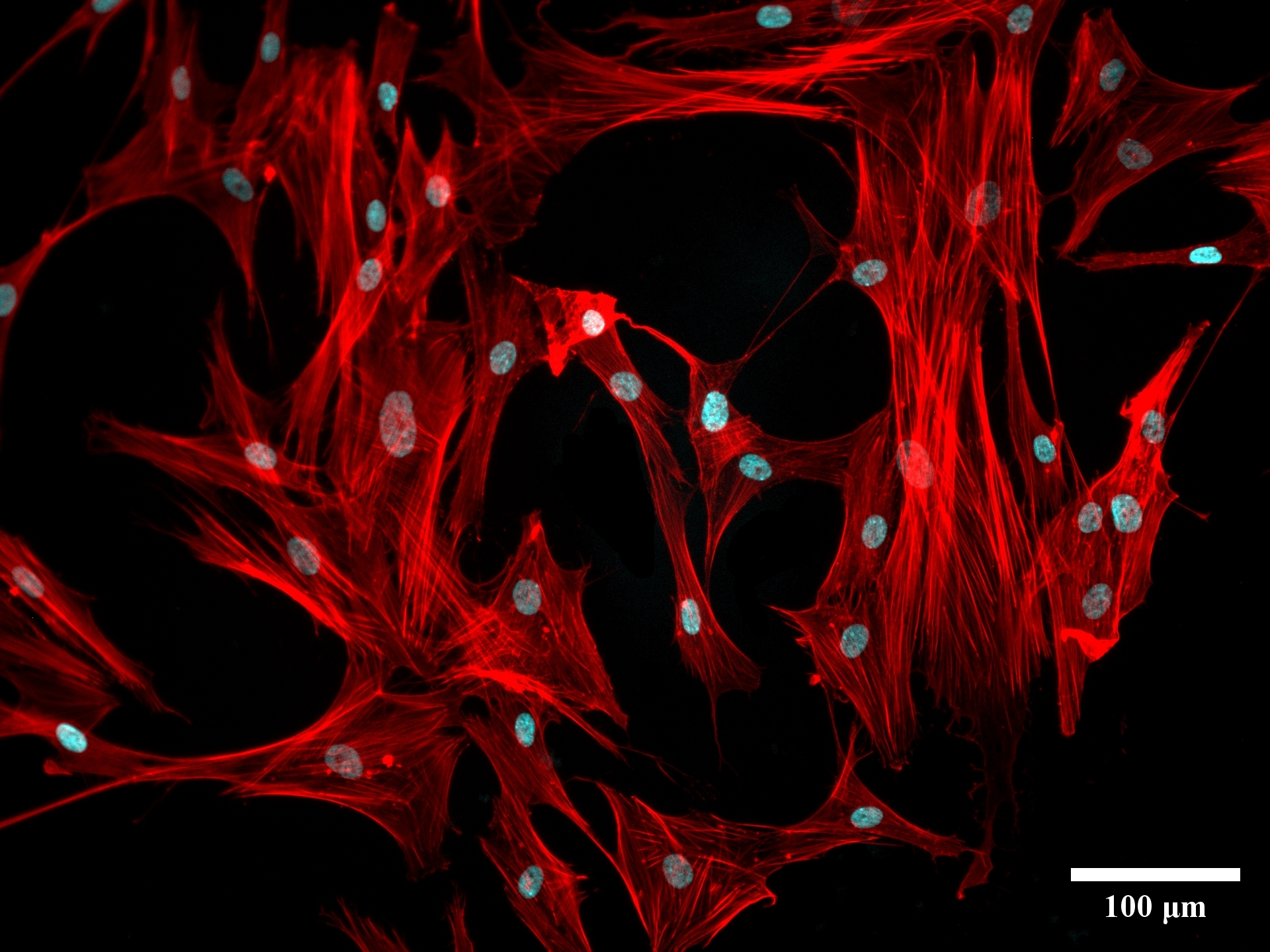
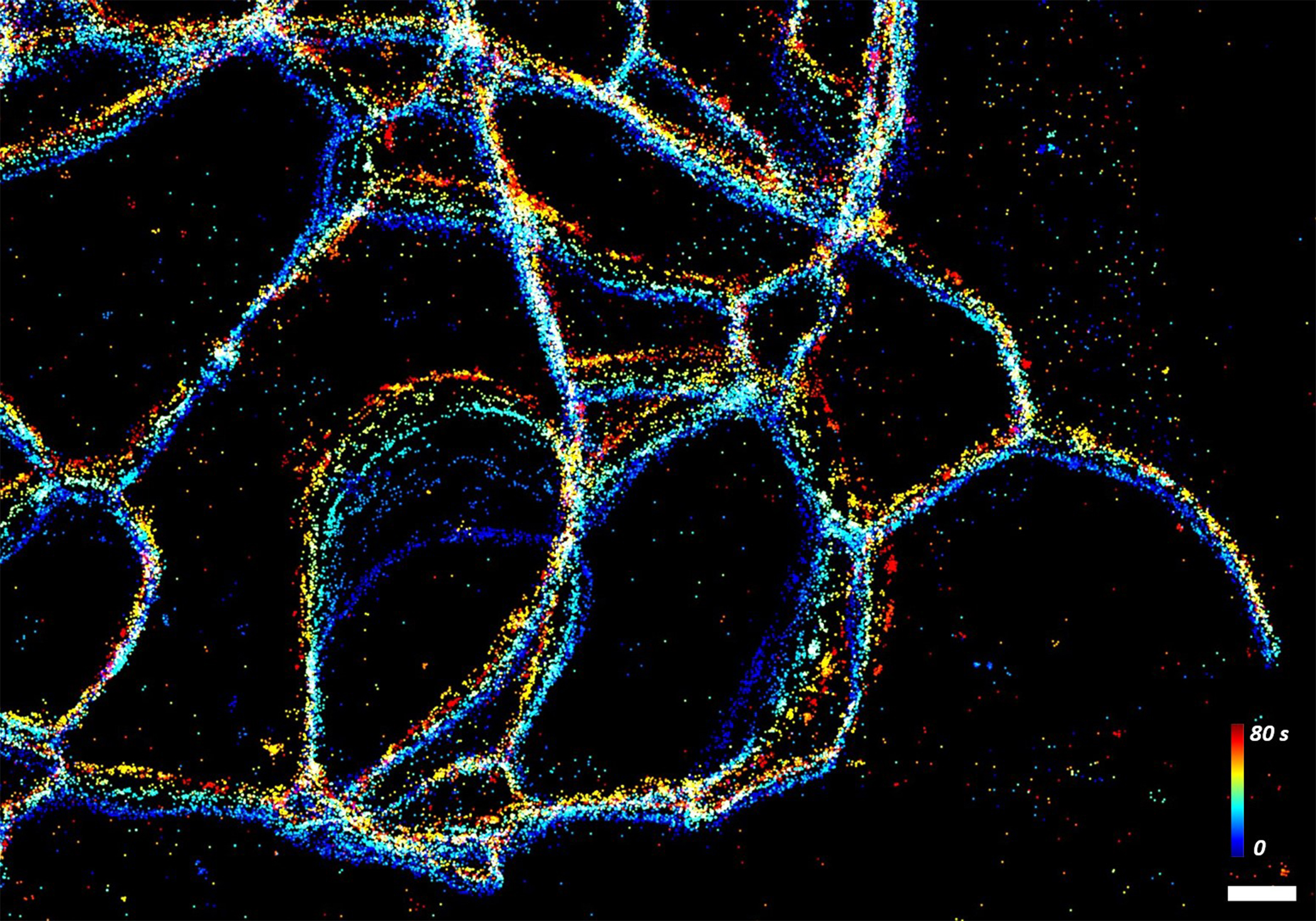
Creation: Microbes from the soil in West Lafayette were cultured on the surface of gelatin-halide film prints.
Caption: This photograph was taken on my 1975 Yashica film camera on a trip to the Galápagos Islands of Ecuador. Prior to inkjet printing in the 1990’s, photographs were printed on gelatin-halide emulsions like these. Taking advantage of microbes’ ability to break down and recycle large proteins, we cultured bacteria and fungi from the soil of West Lafayette on the surface of these prints, and captured their final progress in a moment in time..









-web.jpg)


