Human Betterment Analytics Research Lab - Nan Kong
 Kong's current research involves solving data-driven system optimization problems arising in clinical decision-making, healthcare delivery and public health policy, as well as tackling inherent foundational methodological challenges on data-driven stochastic optimization and model-based reinforcement learning.
Kong's current research involves solving data-driven system optimization problems arising in clinical decision-making, healthcare delivery and public health policy, as well as tackling inherent foundational methodological challenges on data-driven stochastic optimization and model-based reinforcement learning.


 The goal of our research is to understand mechanical properties of cytoskeleton, cells, and tissues via computational models. In addition, we aim to elucidate mechanisms of physiological processes by relating mechanics of the cytoskeleton at subcellular scale to cellular-level and tissue-level mechanics.
The goal of our research is to understand mechanical properties of cytoskeleton, cells, and tissues via computational models. In addition, we aim to elucidate mechanisms of physiological processes by relating mechanics of the cytoskeleton at subcellular scale to cellular-level and tissue-level mechanics.

 The BioCom Laboratory houses a variety of research projects centered around nerve modulation therapies, computational and biophysical modeling.
The BioCom Laboratory houses a variety of research projects centered around nerve modulation therapies, computational and biophysical modeling.  The Pienaar lab uses computational models to understand how pathogens like Ebola virus, HIV and Mycobacteria interact with our immune systems, and how these host-pathogen interactions impact the effectiveness of treatment.
The Pienaar lab uses computational models to understand how pathogens like Ebola virus, HIV and Mycobacteria interact with our immune systems, and how these host-pathogen interactions impact the effectiveness of treatment.
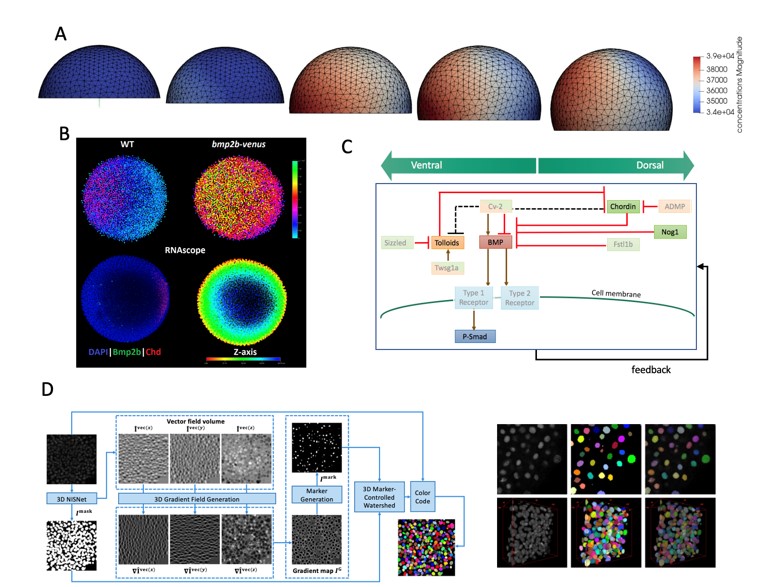
 In the Umulis research group, our focus is to investigate the regulation of signal transduction, and its mechanical and chemical response during development. Specifically, we are interested in elucidating mechanisms of robustness, cell fate decisions and tissue patterning by morphogen gradients.
In the Umulis research group, our focus is to investigate the regulation of signal transduction, and its mechanical and chemical response during development. Specifically, we are interested in elucidating mechanisms of robustness, cell fate decisions and tissue patterning by morphogen gradients. The Translational Medical Image Computing (TMIC) Lab is working at the intersection of medicine and data science, aiming to develop clinically valuable computational tools for patient care.
The Translational Medical Image Computing (TMIC) Lab is working at the intersection of medicine and data science, aiming to develop clinically valuable computational tools for patient care.