3-D-written model to provide better understanding of cancer spread
Previous research has shown that most cancer deaths happen because of how it spreads, or metastasizes, in the body. A major hurdle for treating cancer is not being able to experiment with metastasis itself and knock out what it needs to spread.
Studies in the past have used a 3-D printer to recreate a controlled cancer environment, but these replicas are still not realistic enough for drug screening.
“We need a much finer resolution than what a 3-D printer can create,” said Solorio, an assistant professor of biomedical engineering.
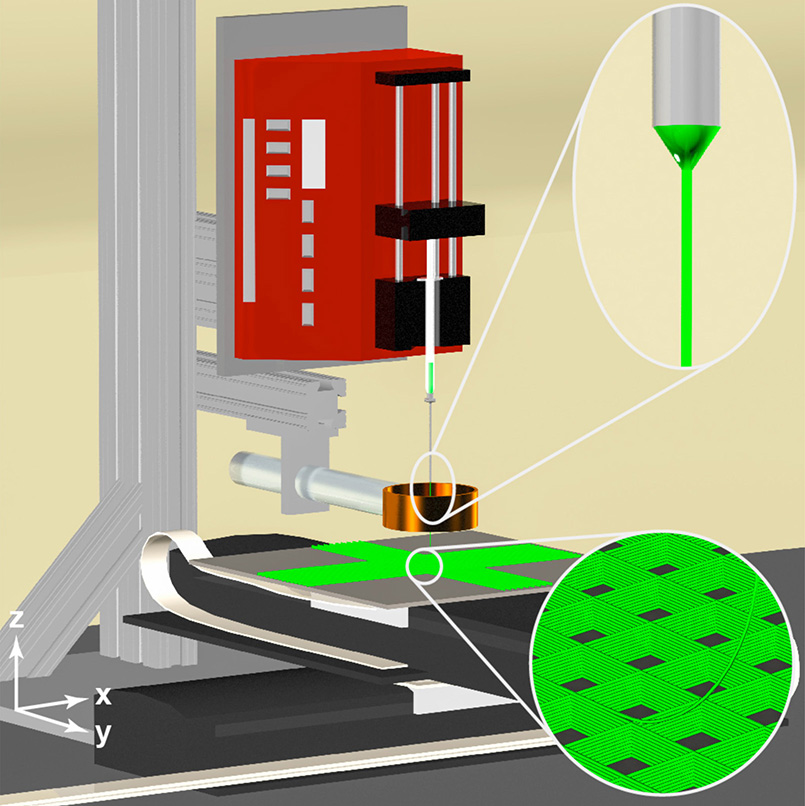
Rather than 3-D printing, Solorio and a team of researchers have proposed 3-D writing. The device that they developed, a 3-D jet writer, acts like a 3-D printer by producing polymer microtissues as they are shaped in the body, but on a smaller, more authentic scale with pore sizes large enough for cells to enter the polymer structure just as they would a system in the body.
3-D jet writing is a fine-tuned form of electrospinning, the process of using a charged syringe containing a polymer solution to draw out a fiber, and then deposit the fiber onto a plate to form a structure. This structure is a scaffold that facilitates cell activity.

Solorio has so far used the device to write a structure that drew in cancer cells to sites in mice where cancer would not normally develop, confirming that the device could create a feasible cancer environment. Solorio’s other studies have increased cancer cells in human samples for better analysis and maintained receptors on these cells that drugs would need to find.
“Ideally, we could use our system as an unbiased drug screening platform where we could screen thousands of compounds, hopefully get data within a week, and get it back to a clinician so that it’s all within a relevant time frame,” Solorio said.
Initial findings published on Feb. 27 in Advanced Materials based on Solorio’s work as part of a team at the University of Michigan Biointerfaces Institute. Continuation of the work is being performed at Purdue and is funded by the National Cancer Institute grant R0019829.
Source: Purdue newsroom
