Microbial and Cellular Pathogens
Microbial and Cellular Pathogens
The BSL-2 Laboratory in LORRE carries out research on detection of food and other pathogens. Detection is based on initial concentration of viable (living) microorganisms using hollow fiber tangential flow filtration (TFF). Pathogens in the recovered concentrate may be detected against a background of non-pathogenic bacteria using PCR, immunoassays, plating on pathogen selective media, imaging, and/or biodynamic (Doppler) imaging.
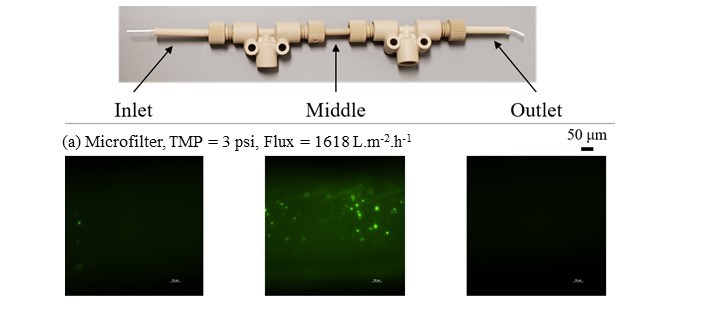
Imaging instruments facilitate monitoring and validation of binding of labeled markers with microbial targets, proteins and peptides, and particulate structures of various sizes. Doppler imaging (developed in the Physics Laboratory of Professor David Nolte: https://www.physics.purdue.edu/index.html) facilitates research on interactions between viral or bacterial pathogens and immortalized mammalian cells. The immortalized cells are grown in our laboratory in the cell culture facility and then characterized using biodynamic imaging (essentially a form of micro-doppler measurement that is able to track the motion of cytoplasm in immortalized cells).

The detection of pathogens is part of a long-term effort of our laboratory and is based on research that has the goal of achieving rapid concentration and recovery of viable (living) microbial populations extracted from plant tissues (vegetables), meats and from other biological fluids. These samples contain both non-pathogenic and pathogenic bacteria. The concentrates are prepared to achieving a sufficiently large population while minimizing the time required to grow the pathogens to detectable levels. This enables probing of these samples for living (viable) pathogenic bacteria in less time than otherwise would otherwise be possible. Detection is based on using various methodologies including immunoassays, plating, PCR, and most recently, direct observation of the effect of living bacteria on living immortalized (mammalian) cell cultures using biodynamic imaging developed by Prof. Nolte’s group. The concentration step itself has been further developed so that a single hollow fiber is able to concentrate 220 mL to 1 mL within 60 min and recover 90% of the microorganisms in the original sample.
Bacteria: Salmonella Enteritidis; Escherichia coli O157 B6-914 (expressing a green fluorescent protein) with both Shiga-toxin genes deleted; Listeria innocua and Listeria monocytogenes. The recombinant microorganism is an E. coli O-157-GFP strain obtained from Dr. Amanda Deering’s laboratory in the Food Science Department at Purdue University. The detailed information for its construction can be found in the Material and Methods section of “Construction and identification of bioluminescent E. coli strains” from Fratamico et al., 1997 (Journal of Food Protection, 60 (10): 1163-1177), and in the Material and Methods section 2.1 from Deering et al., 2011 (Food Research International 45 (2): 1037-1043). Our laboratory collaborates with the laboratory of Professor Eduardo Ximenes in the Department of Environmental and Occupational Health, at Indiana University, Bloomington, IN, with research that is addressing the detection and identification of antibiotic resistant pathogens that cause severe infections in children.
https://publichealth.indiana.edu/research/departments/environmental-occupational-health/index.html
