Artificial Intelligence in Bioinformatics
Species Identification Using Statistical Principal Component Analysis on Genomic Sequence
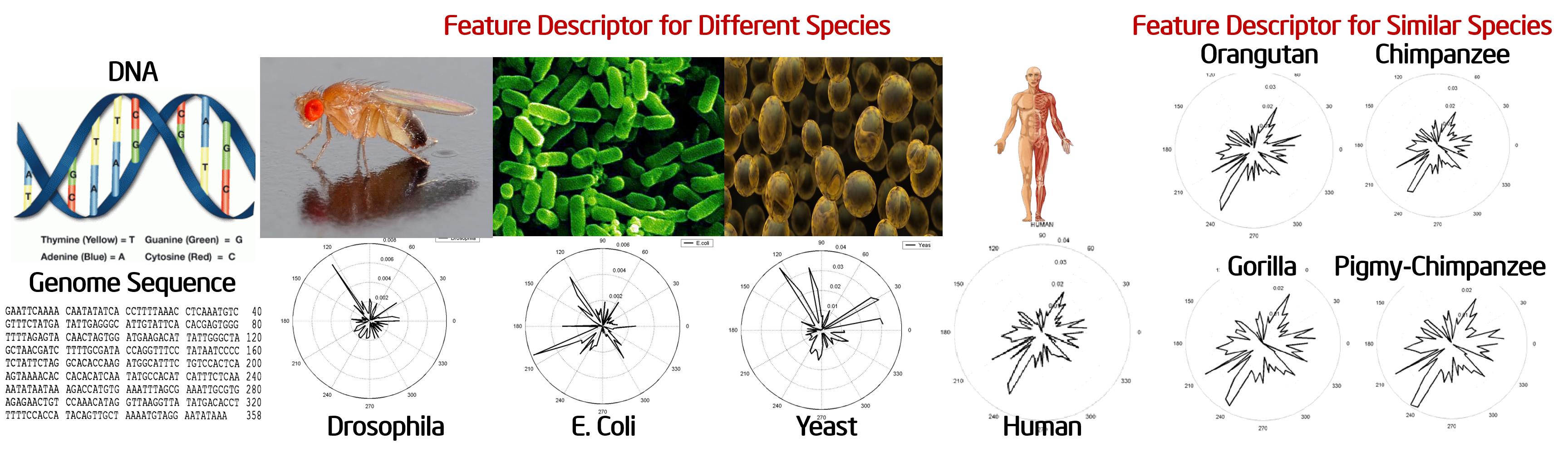
Traditionally, species identification in Bioinformatics is performed using sequence alignment, which is a very compute intensive process. In this work we exploit the statistical similarities and uniqueness of the genomic sequences of different species to enable automatic identification of a species from its genome sequence with significantly less computation. A set of 64 three-tuple keywords is first generated using the four types of bases: A, T, C and G. These keywords are searched on N randomly sampled genome sequences, each of a given length (10,000 elements) and the frequency count for each of the 4^3 =64 keywords is obtained. Principal component analysis is then employed on the frequency counts for N sampled instances. The principal component analysis yields a unique feature descriptor for identifying the species from its genome sequence. The variance of the descriptors for a given genome sequence being negligible, the proposed scheme finds extensive applications in automatic species identification. Using this technique, given a genome sequence, instead of performing sequence alignment, a feature descriptor can be generated to identify the species.
Key Publications : LNCS (Springer) 2005, JCIS 2005