Toward next-generation "super-resolution" 3-D microscopy New achievements in "nanoscope" research attract young investigator funding, promise to reveal dynamics in live cell and tissue specimens.
Toward next-generation "super-resolution" 3-D microscopy
| Subtitle: | New achievements in "nanoscope" research attract young investigator funding, promise to reveal dynamics in live cell and tissue specimens. |
|---|---|
| Magazine Section: | New Faculty Profile |
| Article Type: | Feature |
| Feature CSS: | background-position: 100% 35%; |
| Page CSS: | #article-banner{ background-position : 100% 35% !important; } |
By Emil Venere
Innovations in super-resolution microscopy are promising new capabilities: deeper imaging of tissue samples and live cells, higher resolution of single molecules and, eventually, animal-based studies.
The technology allows researchers to resolve structural and molecular features far smaller than the wavelength of light, representing a powerful and versatile laboratory tool, says Fang Huang, an assistant professor of biomedical engineering at Purdue University.
“The wave nature of light restricts the resolution of conventional light microscopy to about 200 nanometers laterally and 500-700 nanometers in the depth direction, making details of subcellular structures and protein assemblies unresolvable,” he says.

(Source: Photo supplied.)
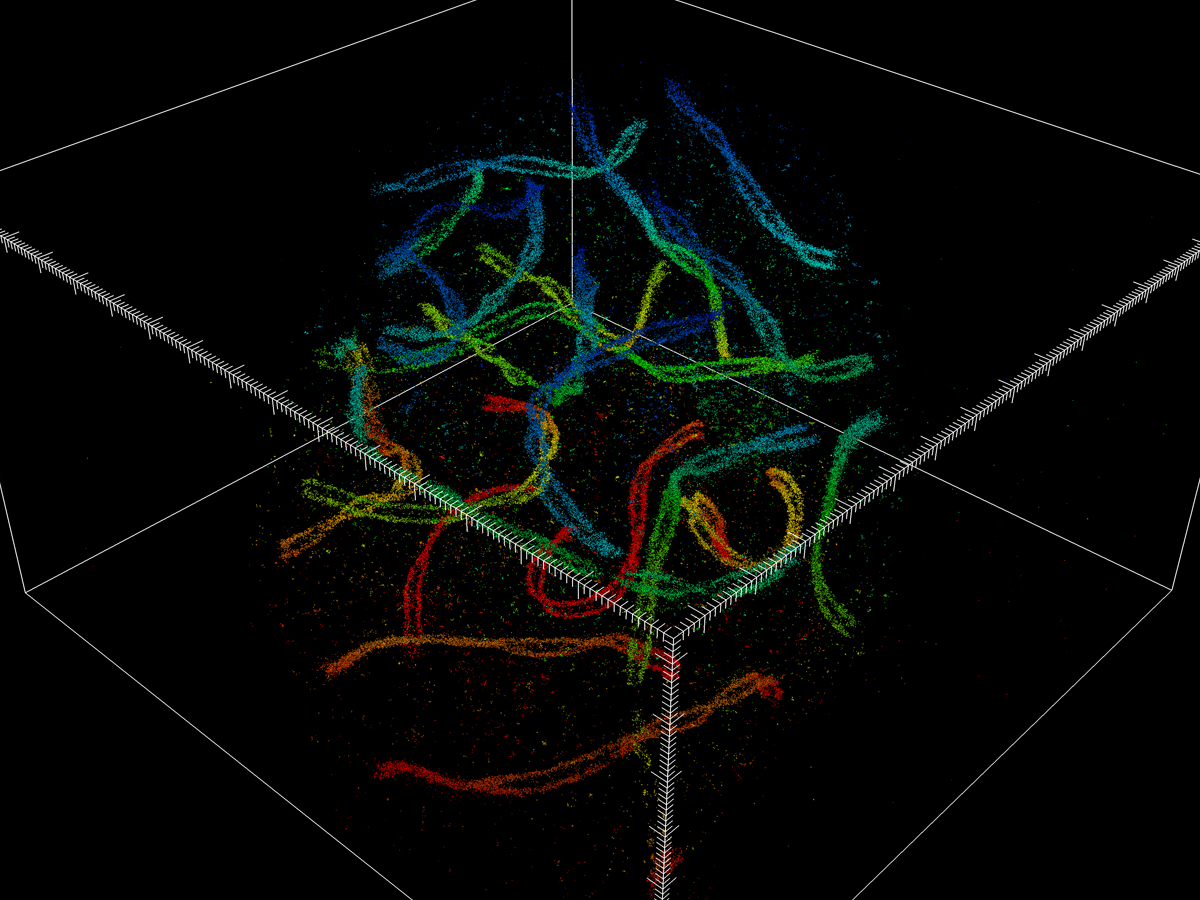
Ultra-high resolution imaging of whole cells
Huang is leading work to develop a next-generation “nanoscope” that promises to reveal dynamic mechanisms inside living cells in unprecedented detail. Molecules inside cells and in structures called organelles can be tagged with either “photo-switchable” fluorescent proteins or organic dyes that are able to glow when exposed to a small amount of ultraviolet light.
“These special fluorescent tags (fluorophores) have two states, on and off,” Huang says. “And you can control the on and off states by shining light on these molecules.”
Super-resolution fluorescence microscopy earned its developers the 2014 Nobel Prize in chemistry and has become an important tool in cell biology research. However, application of this technology has been limited because of challenges in resolving features deep below the surface of samples while maintaining the superior resolution in all three dimensions.
“We have now solved that problem with the development of the new nanoscope, a system called whole-cell 4Pi single-molecule switching nanoscopy (W-4PiSMSN). The approach exploits the wave nature of light by combining every emitted photon from single molecules through two independent but nearly identical optical paths in order to form an interference pattern,” Huang says. “Together with adaptive optics and novel algorithms, precise coordinates of each individual molecule can be derived from its interference patterns even when it is from an organelle deep inside the specimen.”
Pinpointing the coordinates of thousands or millions of molecules makes it possible to reconstruct a 3-D image.
“These results are not pixel-based, but rather a list of single-molecule coordinates,” he says. “You take many camera frames per second, and each frame will capture a different subset of single molecules. So, every dot you see in the reconstruction is one protein that we have labeled. Eventually, there are a lot of points accumulated, and then you can reconstruct the image at super resolution.”
Huang and colleagues’ research findings were detailed in a 2016 paper published in the journal Cell. The paper's co-lead authors were Huang and Yale University postdoctoral research associate George Sirinakis. The team demonstrated the technology by imaging components in live cells, including synaptonemal complexes, which form links between homologous chromosomes; mitochondria and the endoplasmic reticulum, which are critical for cellular functions; the primary cilium, which protrudes from the cell as a sensory antenna; and bacteriophages, which are viruses about 50 nanometers in diameter that infect and kill bacteria.
The nanoscope system allows 3-D imaging of cellular constituents at 10- to 20-nanometer resolution throughout entire mammalian cells, powerful enough to reconstruct the fine features of viruses. Until now, such resolution was only possible using electron microscopy, which requires samples to be treated, killing the cells.
“One goal is to further push the envelope in the direction of live-cell and tissue imaging, the two major challenges for modern super-resolution techniques. We aim to allow visualization of cellular functions live in their physiological conditions at the nanoscale,” Huang says. “Electron microscopy has brought many discoveries but is unable to visualize living processes. We want to see the dynamics in living specimens and use this technology to ultimately understand cytokinesis, the final step in cell division, and to visualize molecular interactions and functions at 1- to 10-nanometer resolution.”

(Photo courtesy of Huang et al./Cell)
Federal funding for breakthrough imaging
The Huang lab recently received two new federal grants: a $1.8 million award from the National Institutes of Health (NIH) and $500,000 from the Defense Advanced Research Projects Agency (DARPA).
“The NIH and DARPA awards facilitate two different research directions in my lab that also are synergistic parts,” Huang says. “I’m hoping we can combine elements of both projects to further push the envelope of super-resolution imaging and to shed light on biological processes that are otherwise hidden under the diffraction limit.”
The NIH Maximizing Investigators’ Research Award (MIRA) for New and Early Stage Investigators is a five-year grant for a project titled “Interferometric 3-D super-resolution imaging and structure and stoichiometry mapping in live cells.”
“The NIH grant will enable us to create a new image modality for building dynamic structural models of large protein complexes in live cells,” says Huang, who came to Purdue in 2015 after postdoctoral training in the Department of Cell Biology at Yale. “These super-resolution systems produce an incredible amount of single-molecule data. It is a challenge and an opportunity for us to explore this new realm and to create new observation tools with this unique resource.”
The DARPA Young Faculty Award will support two years of research with the potential for an additional year to develop a novel nanoscope that significantly improves imaging depth and volume while minimizing light-induced toxicity. The long-range goal is to allow super-resolution studies in live research animals.
Astronomy’s adaptive optics
To achieve this, Huang has harnessed the same kind of adaptive optics used in astronomy — deformable mirrors that change shape to compensate for light distortion. In astronomy, the deformable mirrors in telescopes are used to compensate for atmospheric distortion to yield clear images of celestial objects. Deformable mirrors also can be used to counteract the distortion caused when light passes through biological tissue.
Huang’s system integrates the deformable mirrors in a microscope using two objectives, one above and one below the sample, and introduces a set of new algorithms to pinpoint molecular positions of proteins deep inside cells.
“We use two objectives to collect almost all of the photons emitted from every single molecule that we turn on in the experiment, and we coherently combine them to create an interference pattern of that single molecule fluorescence,” he says. “Originating from a molecular tag on cellular organelles, photons travel through two independent but closely matched interference arms, thanks to adaptive optics, then interfere with themselves to create a standing wave pattern that is extremely sensitive towards the exact position of the single molecule in the cell. This sensitivity translates into a significantly improved resolution compared to conventional super-resolution methods.”

(Photo courtesy of Huang et al./Cell)
Nanoscopy’s multiple disciplines
Huang’s lab combines instrument development with biological sciences, drawing on a range of disciplines, including physics, engineering, signal processing and statistics. “One challenge, as well as an opportunity, is to combine the core expertise from these different fields,” he says. “We also seek to invent algorithms that take advantage of concepts in mathematics, statistics and signal processing. We work closely as a team and also extensively collaborate with biologists around the nation. We are genuinely excited to apply our developed imaging systems to study molecular mechanisms of cell division and to tackle other fundamental biological questions that have profound impact on our understanding of the nature of life.”