Microscopy Image Analysis

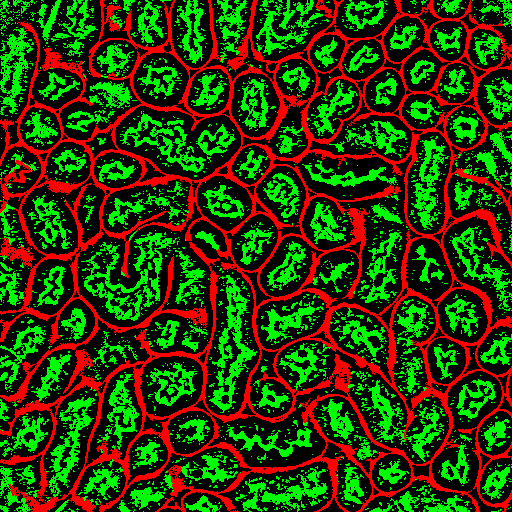
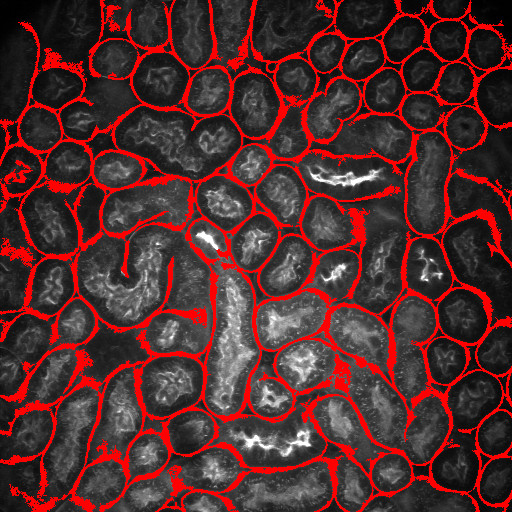
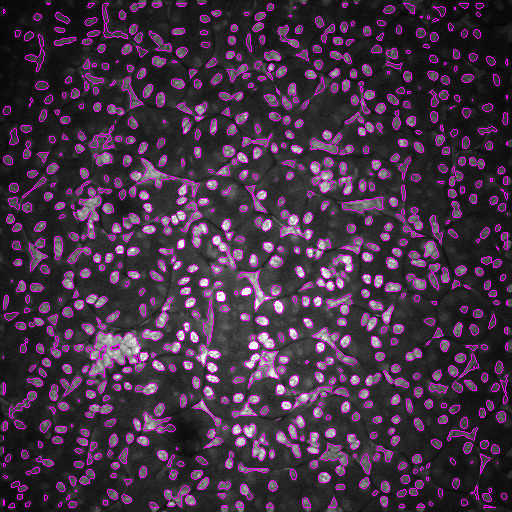

Optical microscopy has become one of the most powerful techniques in biomedical research, particularly when combined with digital image analysis. Since an optical microscope uses visible light and a system of lenses to magnify images, as opposed to electrons and electromagnets present in electron microscopes, the size of the smallest samples an optical microscope is able to resolve is directly proportional to the wavelength of visible light. Among all types of optical microscopes, one variety is a fluorescence microscope, which uses fluorescence instead of reflection and absorption as a source of image contrast.
Optical microscopy exhibits many challenges for digital image analysis. In general, microscopy volumes are inherently anisotropic, suffer from decreasing contrast with tissue depth, lack object edge detail, and characteristically have low signal levels.
Image analysis is motivated by the desire to quantify biological characteristics such as cell count and tissue volume. This project involves the development of methods for an integrated approach to segmentation and registration of intravital microscopy image sets. Image data are acquired using one of two distinct techniques. One data set type consists of a series of images corresponding to focal planes looking deeper in the tissue (three dimensional data), and a second type consists of a series of images corresponding to a sequence of time instances imaging a single focal plane (time-series data). Analysis is done via a combination of segmentation and registration techniques, but is complicated by factors such as live specimen motion during image acquisition. In particular, we are investigating novel methods that utilize image enhancement, spatial filtering, rigid and non-rigid registration, temporal filtering and 2D and 3D segmentation.
Our team consists of researchers from Purdue University (West Lafayette and IUPUI) and the Indiana O’Brien Center for Advanced Microscopic Analysis that is part of the Indiana University School of Medicine, Division of Nephrology.
Our team has developed a tool named Distributed and Networked Analysis of Volumetric Image Data (DINAVID)
that can process and visualize microscopy data via a client-server system.
Link to a description of DINAVID
Link to the DINAVID system
This work is supported by a George M. O’Brien Award from the National Institutes of Health NIH/NIDDK P50 DK 61594.



