Quantitative and Systems Biology Research Group
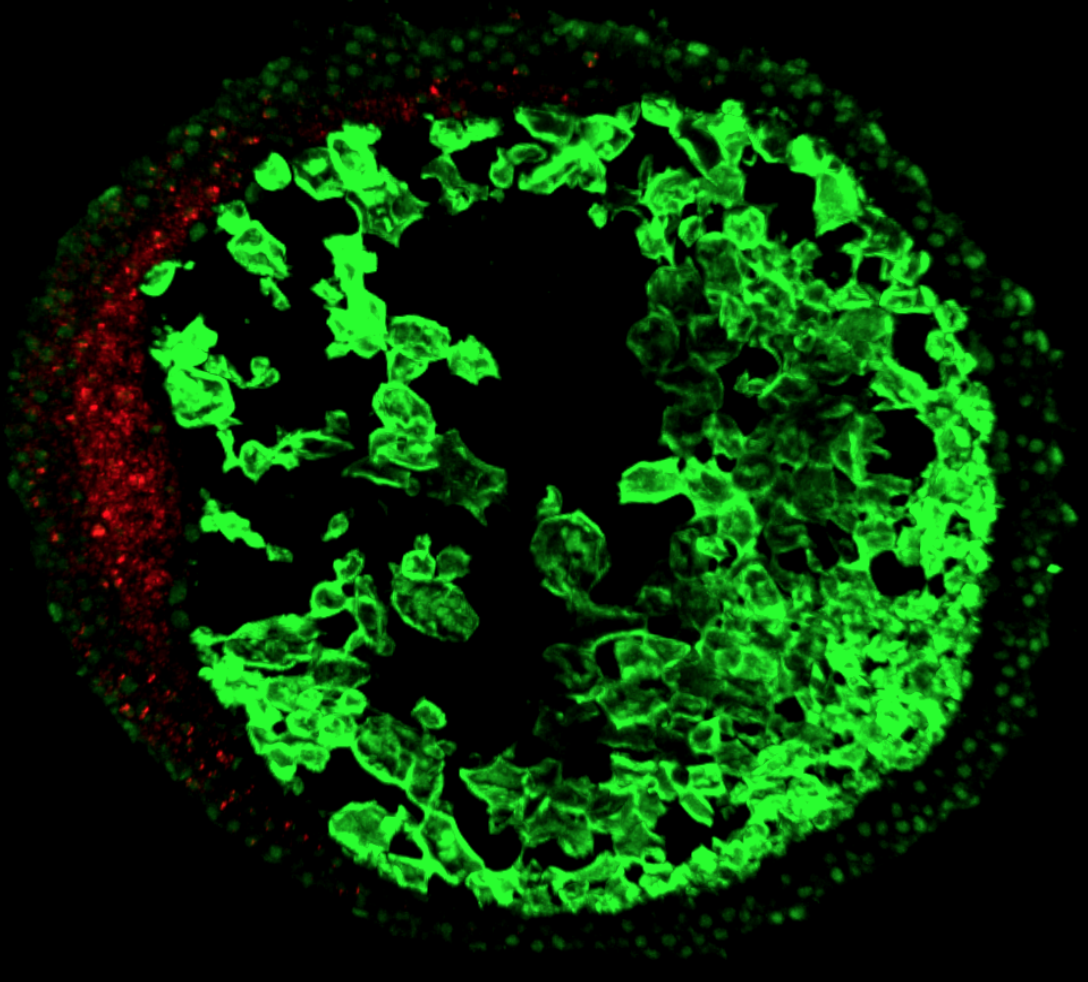
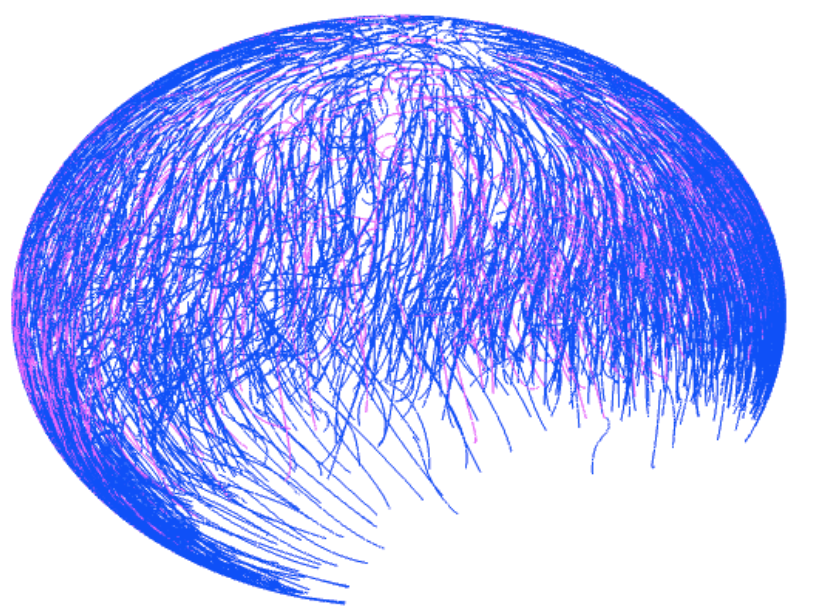
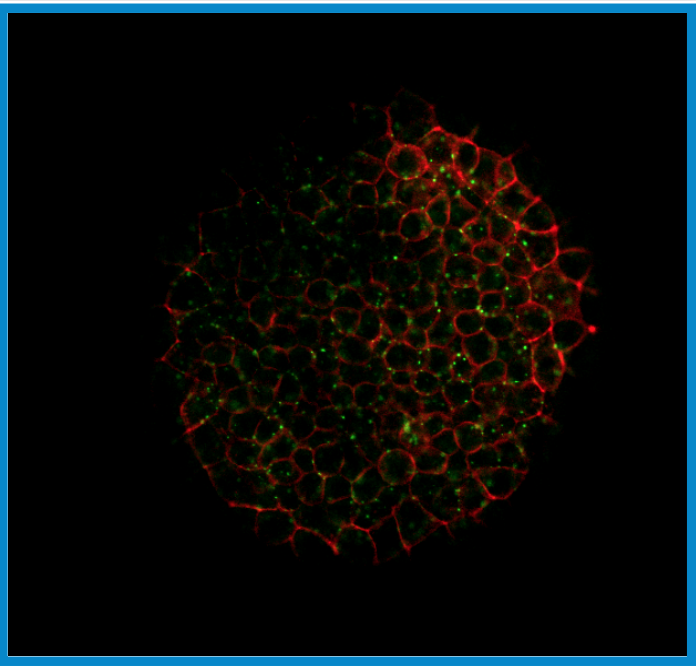
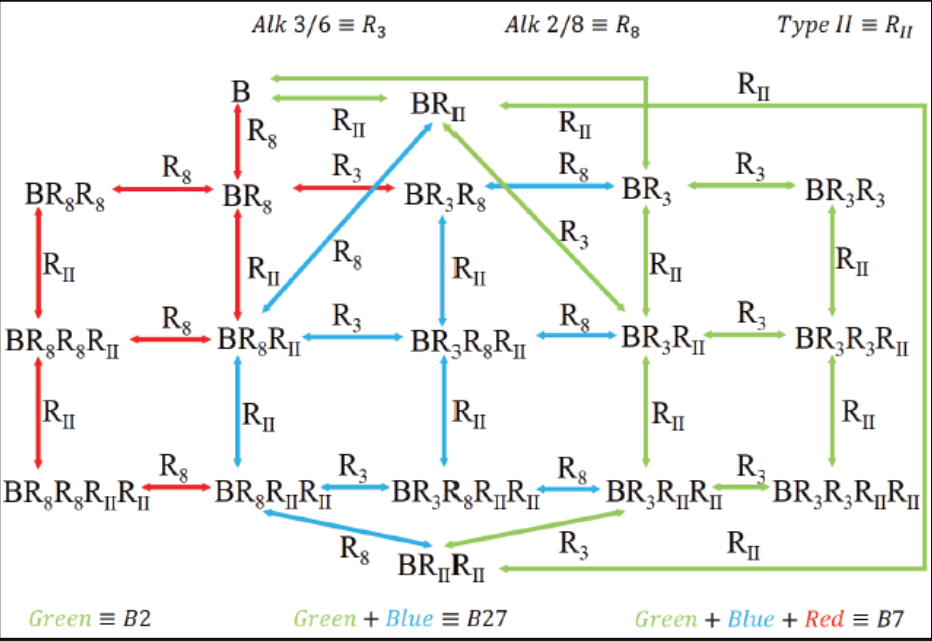
The focus of our research is to investigate the regulation of signal transduction in development. There are three main focuses in our laboratory that can be found under the research tab. The first main topic is modeling, analysis, and discovery of Bone Morphogenetic Protein signaling networks in Drosophila stem cells maintenance and embryo development. The second is developing new methods for data/model integration in biological engineering. Lastly, systems analysis of BMP pattern formation in zebrafish.
Emergent Mechanisms in Biology of Robustness Integration and Organization Institute
Professor Umulis is Director of the Emergent Mechanisms in Biology of Robustness, Integration, and Organization (EMBRIO) Institute. Follow the link below for more details.
https://www.purdue.edu/research/embrio/