Sangtae Kim, Donald W. Feddersen Distinguished Professor Schools of Chemical Engineering and Mechanical Engineering Purdue University
As we approach the second decade of the post-genomic era, the pharmaceutical and biotech R&D worlds appear to be in disarray. The promise of genomics has been replaced largely by wave upon wave of bad news. In 2007 alone, Pfizer announced the closure of its large R&D center in Ann Arbor (where atorvastatin, the #1 drug of all time was discovered) and GlaxoSmithKline announced the termination of eleven projects in the mid-stage (Phase II Clinical Trials) of its R&D pipeline -- devasting what was viewed by many as one of the better R&D pipelines in the industry. And even after reaching the market, breakthrough therapeutic products are recalled as rare and idiosyncratic adverse events (side effects) tip the cost-benefit equation. The problems are industry-wide and not confined to one or two companies -- suggesting that fundamental forces are at play. Our research group is in a special position to combine a high-level industrial perspective with a long track record of imaginative, high-risk academic explorations to break with the unproductive mainstream. The pharmaceutical informatics research thrust area in our group aims to establish new mathematical, informatics and computational foundations to broaden the technological base for harvesting the fruits of genomics.
"Shots on Goal" -- the key to understanding what went wrong and is continuing to go wrong: In conjunction with the emergence of high throughput (massively parallel) technologies, the concept of "shots on goal" carried the day in the mid 90's. Imagine goals scored based on a certain (fixed) percentage of shots passing the goaltender and reaching the target. In such a scenario, of more shots leading to more goals, the obvious strategy is to scale up to large R&D enterprises based on the premise of a high expected return on investment (ROI) for each shot on goal. But what if the net is guarded by a filter and the ball (or puck) is larger than the mesh size? Then it is the loss of shareholder equity that scales with the number of shots on goal. In such a scenario, resources should be directed at widening the mesh; but it is the nature of human societies that those with a vested interest in the goal-shooting infrastructure are unlikely to explore and create the necessary technological disruptions.
Our initial explorations in pharmaceutical informatics are motivated by the near misses -- breakthrough therapeutics for severe diseases that nevertheless are recalled because of serious side effects in a small subset of the population. The nature of the values in our society are such that even highly effective treatments that mitigate the hardships of serious diseases such as MS are recalled if a small subgroup (i.e. one-in-a-thousand) suffer serious consequences. If the subgroup can be identified a priori, warning labels and alerts to physicians will save the day and the product. But more often than not, the reaction is idiosyncratic -- indeed Websters On Line dictionary uses the individual response to drug treatment as the example in the definition of idiosyncratic! But to us, that is simply the challenge for extending our base of scientific capabilities.
Our new thrusts operate on several scales: at the biomolecular level, we are combining the unprecedented availability of high performance computing (HPC) resources with new (in-house) informatics tools to explore cross-reactivity in the space of druggable targets. Combine this with our connections to the systems biology community, we are in a position to motivate large-scale, community-based explorations of the molecular basis of off-target activity ("side effects") in pharmacology.
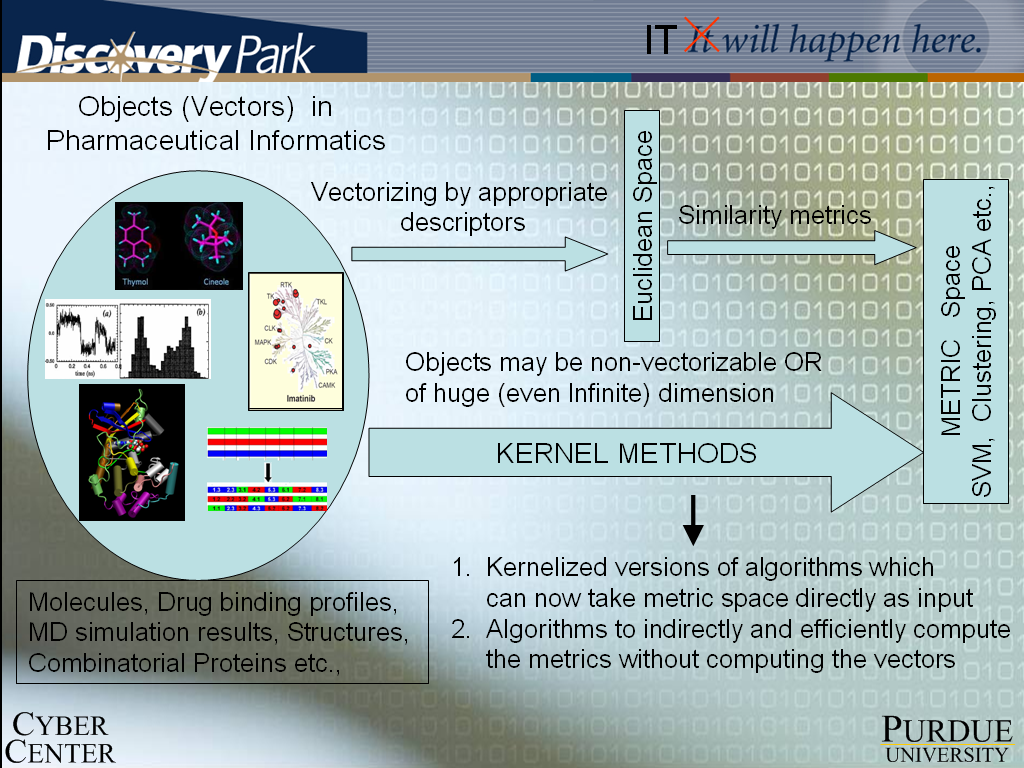
Beyond the molecular level we depend on collaborations with industry visionaries; for we combine our zeal for new advances in informatics and the mathematical sciences with a healthy dose of realism that construction of "useful" metric (feature) spaces -- from a virtually infinite world of possibilities, is unlikely to come about as an exercise in deep-thinking in an ivory tower. This also motivates us to collaborate with experts in dynamic data discovery and mining of R&D data vaults. The early returns are reinforcing our strategic commitment to explore the uncharted territory of kernel methods applied to the appropriate and novel feature spaces; these mathematical constructs depict efficiently key non-pairwise and non-local biomolecular interactions and form the foundational link to the macro-scale of "clinical informatics" and population diversity.
The road to "personalized" medicines runs through the valley of "depersonalized" medicines.

Purdue University, West Lafayette, Indiana
office: 102 Mechanical Engineering
e-mail: kim55@purdue.edu